Putative enhancer sites in the bovine genome are enriched with variants affecting complex traits
Résumé
AbstractBackgroundEnhancers are non-coding DNA sequences, which when they are bound by specific proteins increase the level of gene transcription. Enhancers activate unique gene expression patterns within cells of different types or under different conditions. Enhancers are key contributors to gene regulation, and causative variants that affect quantitative traits in humans and mice have been located in enhancer regions. However, in the bovine genome, enhancers as well as other regulatory elements are not yet well defined. In this paper, we sought to improve the annotation of bovine enhancer regions by using publicly available mammalian enhancer information. To test if the identified putative bovine enhancer regions are enriched with functional variants that affect milk production traits, we performed genome-wide association studies using imputed whole-genome sequence data followed by meta-analysis and enrichment analysis.ResultsWe produced a library of candidate bovine enhancer regions by using publicly available bovine ChIP-Seq enhancer data in combination with enhancer data that were identified based on sequence homology with human and mouse enhancer databases. We found that imputed whole-genome sequence variants associated with milk production traits in 16,581 dairy cattle were enriched with enhancer regions that were marked by bovine-liver H3K4me3 and H3K27ac histone modifications from both permutation tests and gene set enrichment analysis. Enhancer regions that were identified based on sequence homology with human and mouse enhancer regions were not as strongly enriched with trait-associated sequence variants as the bovine ChIP-Seq candidate enhancer regions. The bovine ChIP-Seq enriched enhancer regions were located near genes and quantitative trait loci that are associated with pregnancy, growth, disease resistance, meat quality and quantity, and milk quality and quantity traits in dairy and beef cattle.ConclusionsOur results suggest that sequence variants within enhancer regions that are located in bovine non-coding genomic regions contribute to the variation in complex traits. The level of enrichment was higher in bovine-specific enhancer regions that were identified by detecting histone modifications H3K4me3 and H3K27ac in bovine liver tissues than in enhancer regions identified by sequence homology with human and mouse data. These results highlight the need to use bovine-specific experimental data for the identification of enhancer regions.
Domaines
Sciences du Vivant [q-bio]
Fichier principal
 12711_2017_Article_331.pdf (5.14 Mo)
Télécharger le fichier
12711_2017_Article_331.pdf (5.14 Mo)
Télécharger le fichier
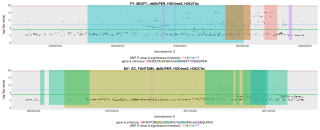 12711_2017_331_MOESM8_ESM.png (2.17 Mo)
Télécharger le fichier
12711_2017_331_MOESM8_ESM.png (2.17 Mo)
Télécharger le fichier
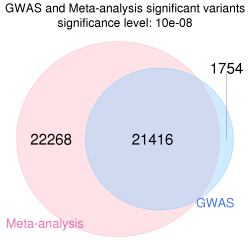 12711_2017_331_MOESM1_ESM.png (115.92 Ko)
Télécharger le fichier
12711_2017_331_MOESM1_ESM.png (115.92 Ko)
Télécharger le fichier
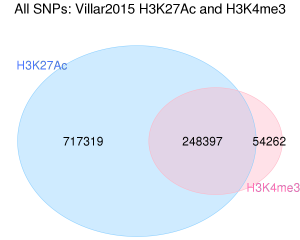 12711_2017_331_MOESM2_ESM.png (90.53 Ko)
Télécharger le fichier
12711_2017_331_MOESM2_ESM.png (90.53 Ko)
Télécharger le fichier
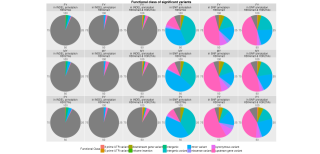 12711_2017_331_MOESM3_ESM.png (819.6 Ko)
Télécharger le fichier
12711_2017_331_MOESM4_ESM.xlsx (11.97 Ko)
Télécharger le fichier
12711_2017_331_MOESM5_ESM.xlsx (10.37 Ko)
Télécharger le fichier
12711_2017_331_MOESM6_ESM.xlsx (10.45 Ko)
Télécharger le fichier
12711_2017_331_MOESM7_ESM.xlsx (12.43 Ko)
Télécharger le fichier
12711_2017_331_MOESM3_ESM.png (819.6 Ko)
Télécharger le fichier
12711_2017_331_MOESM4_ESM.xlsx (11.97 Ko)
Télécharger le fichier
12711_2017_331_MOESM5_ESM.xlsx (10.37 Ko)
Télécharger le fichier
12711_2017_331_MOESM6_ESM.xlsx (10.45 Ko)
Télécharger le fichier
12711_2017_331_MOESM7_ESM.xlsx (12.43 Ko)
Télécharger le fichier
Origine : Publication financée par une institution
Format : Figure, Image
Origine : Fichiers produits par l'(les) auteur(s)
Origine : Fichiers produits par l'(les) auteur(s)
Format : Figure, Image
Origine : Fichiers produits par l'(les) auteur(s)
Origine : Fichiers produits par l'(les) auteur(s)
Format : Figure, Image
Origine : Fichiers produits par l'(les) auteur(s)
Origine : Fichiers produits par l'(les) auteur(s)
Format : Figure, Image
Origine : Fichiers produits par l'(les) auteur(s)
Origine : Fichiers produits par l'(les) auteur(s)
Origine : Fichiers produits par l'(les) auteur(s)
Origine : Fichiers produits par l'(les) auteur(s)
Origine : Fichiers produits par l'(les) auteur(s)
Origine : Fichiers produits par l'(les) auteur(s)
Loading...