| |
| J Viral Hepat. 2006 April; 13(4): 278–88. doi: 10.1111/j.1365-2893.2005.00712.x.A prospective study of the evolution of lamivudine resistance mutations in patients with chronic hepatitis B treated with lamivudine Fabien Zoulim,1* Thierry Poynard,2 Françoise Degos,3 Alain Slama,4 Abdelkhader Al Hasnaoui,4 Patrick Blin,5,6 Florence Mercier,7 Paul Deny,8 Paul Landais,9 Parviz Parvaz,10 and Christian Trépo1 1Virus des hépatites et pathologies associées
INSERM : U271, Université Claude Bernard - Lyon I, Centre de Recherche Inserm
151, Cours Albert Thomas
69424 LYON CEDEX 03,FR 2Service d'hépato-gastro-entérologie
AP-HP, Hôpital de La Pitié-Salpêtrière, Université Pierre et Marie Curie - Paris VI, 47-83, boulevard de l'Hôpital
75651 PARIS Cedex 13,FR 3Service d'hépato-gastro-entérologie
AP-HP, Hôpital Beaujon, Clichy,FR 4GlaxoSmithKline Laboratory
GlaxoSmithKline, Marly-Le-Roi,FR 5Epidemiology department
Icare, Massy,FR 6Département de pharmacologie
Université de Bordeaux, Bordeaux,FR 7STAT-PROCESS
STAT-PROCESS, Paris,FR 8Laboratoire de virologie
AP-HP, Hôpital Avicenne, Université Paris-Nord - Paris XIII, Bobigny,FR 9Service de Biostatistique et d'Informatique Médicale
CHU Necker-Enfants Malades, Paris,FR 10Laboratoire de Virologie
CHU Lyon, Lyon,FR |
Background/Aims Lamivudine resistance has been described in subjects with chronic hepatitis B infections, associated with mutations in the viral polymerase gene. The objective of this study was to estimate the emergence rate of lamivudine-resistant viral strains and their consequences over a two-year period. Methods We evaluated 283 lamivudine-naive subjects with chronic hepatitis B. Clinical and virological features were assessed at inclusion and every six months thereafter. Viral DNA was characterised using PCR-based sequencing. Potential risk factors for the emergence of lamivudine resistance mutations were assessed using logistic regression analysis. Results The annualised incidence rate for viral polymerase mutations was 22%. The only independent risk factor identified was high viral load at inclusion. Detectable viral DNA and elevated transaminases were more frequent in subjects harbouring mutant viral strains, and these underwent a lower rate of HBe seroconversion. All subjects responded favourably to treatment, with no difference in symptoms between the two groups. Conclusions This prospective cohort study identified lamivudine resistant mutations emerging in 22% of subjects yearly, which were apparently not associated with clinical aggravation over the study period. MeSH keywords: Adolescent, Adult, Aged, Aged, 80 and over, Cohort Studies, DNA, Viral, chemistry, genetics, Drug Resistance, Viral, genetics, Female, France, Hepatitis B Antibodies, blood, Hepatitis B e Antigens, blood, Hepatitis B virus, genetics, Hepatitis B, Chronic, drug therapy, virology, Humans, Lamivudine, therapeutic use, Male, Middle Aged, Mutation, Polymerase Chain Reaction, Prospective Studies, Reverse Transcriptase Inhibitors, therapeutic use, Sequence Analysis, DNA, Statistics, Nonparametric Author keywords: Chronic hepatitis, hepatitis B virus, antivirals, drug resistance |
Chronic hepatitis B infections remain a major public health challenge (1). The introduction of lamivudine represented a significant advance in the treatment of these infections. This drug is a nucleoside analogue that prevents viral replication by inhibiting the viral reverse transcriptase activity (2). Lamivudine has been demonstrated to stimulate viral clearance and improve clinical status in several randomised clinical trials (3–5), and to provide sustained benefit over periods up to four years (6, 7), including in subjects infected with HBe-Ag negative viral strains (8). The efficacy of lamivudine is, however, compromised by the development of viral resistance (9–11), due to the selection of HBV variants containing mutations in the YMDD motif of the hepatitis B polymerase (12, 13). The long-term clinical consequences of these lamivudine-resistant mutants are poorly understood. Most of the available data has come from analyses of subjects included in randomised clinical trials. As these are a highly selected population, notably in terms of comorbidities, the extent to which these findings can be generalised to the general population is not clear. In an analysis of data from four randomised clinical trials (14), viral breakthrough and flares of transaminase activity occurred frequently but these were less intense than pre-treatment disease activity. Clinically significant symptoms of re-emergent liver disease have been sometimes reported, involving severe complications such as hepatic decompensation (15). More recently, data has become available from long-term extensions of two of these trials. One of these, in patients with HBe-Ag negative hepatitis B, performed liver biopsies before and three to four years after initiation of lamivudine treatment (16). Development of resistance mutations was associated with histological evidence for worsening of hepatic disease in certain patients. The other study (17) concluded that improvement in terms of seroconversion and transaminase activity after four years could be observed in spite of the emergence of resistant strains bearing polymerase mutations. In patients with cirrhosis or advanced fibrosis, a five-year randomised placebo-controlled trial of lamivudine revealed a mutation rate of 49% after a mean treatment duration of 32.4 months (18). The proportion of patients whose Child-Pugh score increased was higher (7%) in those patients developing mutations than in those in whom it did not (1%). A retrospective analysis of patients infected with HBe-Ag negative HBV strains, many of whom had severe liver disease at inclusion, found that viral breakthrough was associated with a higher probability of clinical deterioration (19). Some small observational studies have been performed in subjects starting lamivudine treatment (20–22), and these have yielded similar resistance emergence rates to analyses of clinical trials. To identify more precisely the extent and impact of lamivudine-resistant mutants in unselected subjects with chronic hepatitis B infections under standard conditions of care, we have carried out a large prospective pharmacoepidemiological study of HBV polymerase mutants in a consecutive cohort of subjects with chronic hepatitis B infections treated in specialist hepatology centres in France. The principal objective of the study was to estimate the annual incidence of these mutants following initiation of lamivudine therapy over a two-year follow-up period. Secondary objectives included assessment of the clinical course of infection following emergence of mutations and determination of clinical and biological factors predictive of lamivudine resistance. |
This prospective cohort study was performed in fifty-two hospital Hepato-Gastroenterology or Internal Medicine departments across France. Subject selection Each participating centre included the next six subjects over the age of eighteen who presented with an active chronic hepatitis B infection for which the investigator decided to initiate lamivudine treatment. Exclusion criteria included previous lamivudine exposure, dialysis, HIV infection, immunosuppressant chemotherapy or receipt of an organ graft. Lamivudine was only provided if the conditions set out in the prescribing information for the drug were fulfilled, notably concerning pregnancy. Study design Subjects were followed up every six months for 24 months after starting lamivudine treatment. At baseline, data was recorded on sociodemographic characteristics, alcohol consumption, risk factors for hepatitis B infections, other viral infections, duration of hepatitis B infection, symptom presentation, results of previous serological tests or liver biopsies and previous treatments for hepatitis B. Histological abnormalities were scored in biopsy samples using Knodell ( 23) and Metavir scales ( 24). In addition, symptomatology was described, the results of any biological tests assessed and the lamivudine treatment schedule evaluated. A blood sample was taken at each study visit for virological characterisation, detection of HBe antigen and anti-HBe antibodies, and measurement of transaminase levels. Virology Serum HBe antigens and antibodies were measured using routine procedures in each centre. Seroconversion was defined as the loss of HBe antigen concomitant with the appearance of HBe antibodies. Blood samples were analysed for viral genome centrally in a blinded fashion with respect to the clinical data. Viral DNA was extracted from serum using the QIAmp DNA Blood Mini Kit (QIAGEN, Courtaboeuf, France). Baseline HBV DNA was quantified using the Versant branched DNA kit version 3.0 (Bayer, France) with a lower limit of detection of 357 IU/mL (one IU/mL represents 5.6 copies/mL). The HBV polymerase gene was sequenced following amplification by polymerase chain reaction (PCR) using appropriate oligonucleotide primers. The lower limit of detection of the PCR assay was 70 IU/mL. The sequencing reaction was performed using labelled nested primers (CY5.5-POL3M and CY5.0-P4M), CY5/CY5.5- Dye Primer Kit, Long-Read TOWER sequencer and Opengene System software (Visible Genetics, Evry, France). INNO-LiPA genotyping and PreCore mutant detection kits were used as described in more detail elsewhere (25). Statistical analysis The study population included all eligible subjects who provided an exploitable blood sample at baseline and at least once during the follow-up period. When follow-up analyses were missing, mutation status was affected to the missing time points by the principle of last observation carried forward. Categorical variables were compared with the χ 2 test or Fisher’s exact test and quantitative variables with the Kruskal-Wallis test. Determinants of mutations were assessed from Odds Ratios calculated using univariate or multivariate logistic regression analysis. Ethics The study was conducted in accordance with the Declaration of Helsinki and pertinent regulatory requirements. Written informed consent was obtained from each subject before inclusion. The protocol was approved by the Ethics Committee of CCPPRB Lyon A. |
Patients included Between October 1999 and March 2001, 298 subjects were included in 52 centres. Fifteen (5.1%) were excluded from the analysis, principally due to the absence of follow-up data on mutation status. In all, 274 subjects (96.8% of the analysable population of 283 subjects and 91.9% of the included population) completed the 24-month treatment period. Sociodemographic, virological and clinical features of included subjects Subjects were subdivided by HBe serotype into 164 HBe-negative subjects (58.0%) and 119 HBe-positive subjects (42.0%). The Sociodemographic and clinical characteristics of the subjects included in the study according to HBe serotype are presented in Table 1. The HBe-negative subjects were on average 7.5 years older than HBe-positive subjects. A large proportion of subjects came from immigrant communities, notably the Mediterranean basin, Asia and Africa. The most frequently encountered clinical symptom was asthenia. Eight subjects had jaundice at inclusion, 24 hepatomegaly and six ascites. Biopsy-confirmed cirrhosis was present in 58 subjects. Transaminase levels were elevated in 86% of subjects, although elevations did not usually exceed three times the upper limit of normal (Table 2). Elevated transaminase levels were more common in HBe-positive subjects. Higher Knodell and Metavir scores were more frequently encountered in HBe-negative subjects (Table 2). Viral load was significantly higher in HBe-positive subjects (Table 2). Mutations in the basic core promoter (BCP) region and the pre-core region were identified in the majority of subjects (84.6%) at inclusion. There was a slight preponderance of promoter mutations compared to stop codon mutations, with both mutations being observed in 35.2% of subjects. BCP mutations were found in both HBeAg positive and negative patients, while pre-core mutations were found mainly in the HBeAg negative population. The correlation between the prevalence of BCP/pre-core mutations obtained from genotyping and HBe serotype was incomplete (Table 2): strains harbouring BCP/pre-core mutations could be isolated from 74.4% of HBe-positive subjects, whereas in twelve HBe-negative subjects (7.7%), no mutations could be detected in either the BCP or the pre-core region. Genotype D was the most frequent HBV genotype followed by genotype A whereas genotypes F and G were very rare (Table 2). Genotype D was twice as frequent in HBe-negative subjects, whilst the opposite distribution was observed for genotypes B and C. At the time of inclusion, 54.4% of the population was treatment-naive; the rest had received predominantly interferon α (data not shown). Only twenty subjects had received previous nucleoside analogue antiviral drugs, principally vidarabine (twelve subjects) and/or famciclovir (ten subjects). Evolution of HBV polymerase gene and BCP/pre-core region over the study duration Over the two-year study, mutations in the YMDD motif of the viral polymerase were observed in 114 subjects, corresponding to an incidence rate of 41.5% (95% confidence limits 35.6% and 47.3%). The time course of emergence of mutations was linear ( Figure 1, panel A). The most frequently observed mutations were L180M, M204V and M204I ( Table 3). In the majority of subjects with mutations, more than one point mutation was identified. The most frequent association was L180M/M204V observed in 48 subjects (42.1% of all subjects with mutations) at the study end, while the M204I variant was mainly observed as a single mutant ( Figure 1, panel B). There was no apparent difference in the rate of emergence of the individual mutations, nor of single versus multiple mutants ( Figure 1, panel B). There was no relationship between emergence of polymerase mutations and either HBe serotype ( Table 3) or BCP/pre-core mutation status (data not shown). HBV strains carrying M204I or M204V mutations were respectively over- and under-represented in subjects carrying strains with a D genotype ( Table 3). Determinants of emergence of lamivudine-resistant mutations Potential determinants of emergence of lamivudine-resistant mutations were assessed in terms of odds ratios. In a first step, baseline demographic, clinical and virological variables were evaluated independently in a univariate model. The significant variables identified in this analysis are presented in Table 4. Age and place of birth were the only demographic variables associated with the emergence of YMDD mutations, which were more frequent in older subjects. Mutations appeared less frequently in subjects of Asian origin, and interestingly genotype C (which is prevalent in Asia) was associated with a lower frequency of lamivudine resistance ( p = 0.013, χ 2 test). Apart from this, there was no other association with viral genotype ( p = 0.090, Fisher’s Exact test). Moreover, the emergence of mutations was not dependent on the presence of BCP/pre-core mutations in the viral populations at inclusion ( p = 0.117, χ 2 test) or of an HBe-negative serotype ( p = 0.318, χ 2 test). For clinical symptoms and risk factors, no significant associations with emergence of YMDD mutations were observed with the exception of Metavir score, with subjects with more pronounced liver pathology at baseline being associated with a higher risk of emergence of mutations. Alcohol consumption, high viral load at inclusion (> 5 × 10 6 IU/ml) and previous drug treatment for hepatitis B infections were also all associated with the development of mutations in the YMDD motif. In a subsequent step, these variables identified in the univariate analysis were entered into a multivariate model. The only such variable to be retained with a significant association with polymerase mutations was viral load. Virological and clinical outcome The proportion of subjects with detectable viral DNA by PCR fell to 39.9% during the first six months after initiation of lamivudine treatment. In subjects in whom lamivudine-resistant strains did not develop, this proportion remained relatively stable and was 34.0% at study end ( Figure 2). On the other hand, in subjects with lamivudine-resistant strains, serum HBV DNA was detectable in 72.6% of subjects at the study end. The difference in presence of serum HBV DNA according to polymerase mutation status was significantly different ( p < 0.001) at 12, 18 and 24 months after initiation of lamivudine treatment. During the first six months of treatment, transaminases remained normal or normalised in 95.7% of the subjects. In subjects without lamivudine-resistant strains, the proportion of subjects with elevated transaminase levels remained below 7.5% throughout the study (Figure 2). In the subjects with lamivudine-resistant strains, the proportion with elevated transaminase levels was significantly higher (between 10% and 15%) at 12, 18 and 24 months. However, these elevations in transaminase levels were not accompanied by liver failure assessed by prothrombin time, or by jaundice. HBe seroconversion between inclusion and study end was assessed in 116 of the 119 subjects who were HBe-positive at baseline. In the 51 such subjects who developed lamivudine-resistant strains, ten seroconverted during the study (19.6%). On the other hand, 26 of the 65 HBe-positive subjects without lamivudine resistant strains had seroconverted after 24 months (40.0%). The difference in the rate of seroconversion between YMDD mutant-positive and YMDD mutant-negative subjects was statistically significant at 18 and 24 months (Figure 2). After 24 months, the rate of seroconversion was higher in subjects with a BCP mutation at baseline (22 subjects: 33.3%) than in those without (13 subjects: 26.5%), although this difference was not statistically significant. In all but four cases (two with YMDD mutations and two without), HBe seroconversion was associated with normalisation of transaminase levels at twenty-four months. Six subjects died during the course of the study. None of the deaths were considered by the investigators to be related to lamivudine treatment. The causes of death were hepatocarcinoma in two cases, hepatocellular failure in two cases (one with previous hepatomegaly and ascites, resolved at the last available estimation, and the other with no prior symptoms reported in the study), leukaemia in one case and one non-determined cause of death. One case with hepatocarcinoma had developed a lamivudine-resistant strain while such a mutant was not detected in the other five cases. Following initiation of treatment, jaundice resolved in the first six months in seven subjects and between six and twelve months in the remaining subject. No cases of jaundice emerged during the study. Ascites resolved in the first six months in five subjects and between six and twelve months in the remaining subject. Two case of ascites emerged during the study, one during the first six months in a subject who subsequently died of hepatocellular failure and one between 12 and 18 months in a subject from whom a YMDD mutant strain had been isolated after six months of treatment, which subsequently resolved. Hepatomegaly disappeared progressively but more slowly, and was still present in two YMDD mutant-positive subjects and four YMDD mutant-negative subjects at study end. In these two groups, hepatomegaly emerged during the study in four subjects and one subject respectively who did not present this symptom at inclusion and recurred after transient resolution in an additional two and one subject respectively. In all but three subjects, these cases of emergent or recurrent hepatomegaly observed during lamivudine treatment had resolved by the next six-monthly evaluation. |
This large, two-year prospective multicentre cohort study evaluated the emergence of lamivudine resistance mutations in 283 subjects with chronic hepatitis B infections starting lamivudine therapy, typical of those treated in specialist hepatology units in France. The subjects were predominantly male and the majority (58.8%) were born outside France, principally in other countries of the Mediterranean basin, Asia and Africa. Significant elevations of liver transaminases (> 3 × ULN) as well as clinical symptoms were only observed for a minority of subjects, although most presented fibrosis or cirrhosis on liver biopsy and had elevated Knodell and Metavir scores. This suggests that hepatitis B infections may often be clinically silent in spite of ongoing liver disease. Therefore, a more active screening of high-risk groups may be useful to identify such silent infections early to prevent complications of the disease. A large number of subjects who were actually infected with a BCP mutant strain did not present an HBe-Ag negative serotype. In these subjects, there was a suggestion that the presence of such a mutation may favour future seroconversion, and this merits exploration in further studies. This mismatch was already observed in the original description of the genotyping methodology used in this study (25) and has since been described in a large national survey of hepatitis B performed in the United States (26) as well as in East Asian populations (27). A potential explanation for this finding is that BCP mutations may attenuate rather than abrogate completely synthesis of RNA encoding the HBe antigen (28). In some cases, loss of HBe antigen following appearance of a BCP mutation may be delayed or incomplete. There is some evidence that the extent of HBe antigen production from viral genome bearing BCP mutations may vary between strains (28), perhaps related to HBV genotype (27). Nonetheless, the mismatch between genotyping and serotyping for BCP mutations suggests that classical HBe serotyping underestimates the prevalence of pre-core mutations and that monitoring of the subjects using genotyping technology may be warranted (25–28) We identified an incidence rate for mutations in the YMDD motif of the HBV polymerase gene of 22.3% over one year and of 41.5% over two years in these subjects starting lamivudine therapy. These data are very similar to those obtained from the analysis of pooled data from four clinical trials reported by Lai et al. (14), who reported a one-year incidence rate of 24% and a two-year rate of 42%. In contrast with the Phase III trials, which imposed rigorous inclusion and exclusion criteria, our study recruited a random sample of subjects with hepatitis B infections fulfilling broad inclusion criteria presenting at specialist hepatology departments in France. The incidence for lamivudine-resistant mutations found in our study may thus reflect the incidence rate in the general population in France. Interestingly, the study of the profile of mutations in the polymerase gene and their kinetics of emergence showed that the M204V mutant is most often found in association with the compensatory L180M mutation, while the M204I mutant is more rarely found in association with this mutation (Figure 1, panel B). The M204I mutant was also found principally in association with the HBV genotype D. These findings may have important clinical implications with respect to the development of new antiviral agents that may present some level of cross-resistance with lamivudine (29, 30). We also attempted to determine features that could predict the emergence of lamivudine-resistant mutations. Initially, a univariate analysis was performed using all the demographic, clinical and virological parameters recorded at inclusion. This identified age >50 years, alcohol consumption, high viral load, high Metavir score and previous antiviral treatment as being associated with higher incidence of polymerase mutation, and Asian origin and C genotype as being associated with lower incidence. However, when entered into a multivariate analysis only viral load was retained as a determinant of emergence of lamivudine-resistant mutations. Nevertheless, the relatively low number of subjects in whom strains with these mutations were found may preclude the identification of clinically relevant determinants with more subtle effects. Other studies such as the one reported by Benhamou et al. (31), did not identify baseline predictive factors for polymerase mutations in 66 subjects co-infected with hepatitis B and HIV treated with lamivudine for four years. Furthermore, our study did not replicate data from randomised clinical trials (14), in which high body mass index, male gender and more severe hepatic disease were found to be predictors of polymerase mutations in multivariate analysis. In small studies, it has been suggested that HBe antigen negativity may be associated with a lower rate of emergence of HBV polymerase mutants during lamivudine treatment (21,32,33). However, our study, in which more patients were enrolled, did not identify a relationship between either HBe serotype or the presence of BCP/pre-core mutations and the emergence of lamivudine resistance mutations over the two-year study period. Another small study has reported that HBV genotypes might be associated with a differential risk of development of lamivudine resistance (34). In contrast, our study found no obvious relationship with the emergence of lamivudine resistance mutations, in agreement with previous smaller scale studies from Japan and France (33, 35). All subjects responded to lamivudine treatment by a reduction of viral load, accompanied by a normalisation of transaminases in the majority of subjects. However, both viral load and transaminases were significantly higher in subjects in whom lamivudine-resistant strains had been identified, suggesting that these mutations are associated with some degree of viral breakthrough and hepatic impact. In our study, a significant difference was observed between the subjects with and without lamivudine-resistant strains in the extent of HBe seroconversion after two years, which was observed in 40.0% of subjects who did not develop mutations in the YMDD motif compared to 19.6% in those subjects who did not. This finding is again consistent with the clinical trial data of Lai et al. (14), who reported one-year seroconversion rates of 21% and 8% for subjects with and without lamivudine-resistant strains respectively. In terms of clinical outcome, a progressive decrease in symptoms of liver disease during the study period was observed. This desirable therapeutic response was observed independently of the presence of lamivudine-resistant HBV strains. There was no difference in the rate of disappearance of symptoms, the frequency of symptoms during the study or in the incidence of new symptoms between subjects with or without these strains. The extent to which emergence of lamivudine-resistant HBV strains is a risk factor for progression of cirrhosis or emergence of fulminant liver disease is not clear. Some studies have found little evidence for more liver damage in those subjects who developed lamivudine-resistance HBV strains (17, 36, 37), whereas many others describe ALT flares, hepatic decompensation, or worsening histology following emergence of lamivudine resistance and viral breakthrough (16, 18, 19, 32, 33, 38, 39). Part of this discrepancy may be related to patient characteristics, with those with severe liver disease or a pre-core mutant infection being more at risk for aggravation in the case of development of lamivudine resistance. In our cohort study, no systematic assessment of histological progression by liver biopsy was performed. However, liver function was assessed in all patients using the non-invasive FibroTest-ActiTest (FT-AT), that has been validated as a surrogate marker for histologically-confirmed liver damage in patients with hepatitis B (40). These data, published elsewhere (41), show that lamivudine treatment led to a significant decrease in necroinflammatory activity and fibrosis. Both the extent of improvement and the proportion of patients in whom improvement was observed were similar in patients with or without lamivudine-resistant HBV strains. However, deterioration was observed in two patients with cirrhosis when treatment was initiated, in both of whom a YMDD-mutant HBV strain was isolated. In conclusion, our cohort study provides new information on the clinical and virological characteristics of subjects starting lamivudine therapy in France. This study revealed an annualised incidence rate for lamivudine resistance mutations in HBV strains isolated from subjects treated with lamivudine in the general population (22%) comparable with previous estimates obtained from clinical trials. The emergence of mutations was apparently not associated with aggravation of the clinical state of the subject, at least during the study period. The only independent variable predictive of emergence of lamivudine resistance mutations was viral load at initiation of treatment. |
The lamivir study group (participating centers): A. Abergel (Clermont-Ferrand), J.P. Arpurt (Avignon), E. Beaujard (Alençon), P.M. Bernard (Bordeaux), R.M. Bory (Lyon), D. Botta-Fridlund (Marseille), M. Bourlière (Marseille), O. Boutet (Bagnols/Cèze), J.P. Bronowicki (Vandoeuvre-Les-Nancy), C. Buffet (Le Kremlin Bicêtre), J.F. Cadranel (Creil), X. Causse (Orléans), P. Chamouard (Strasbourg), P. Couzigou (Pessac), M.T. Dao (Caen), J.M. Debonne (Marseille), F. Degos (Clichy), J. Doll (Le Chesnay), T. Fontanges (Bourgoin Jallieu), N. Ganne (Bondy), J.L Gaudin (Lyon), O. Goria (Rouen), J. Gournay (Nantes), J.D. Grangé (Paris), A. Guillygomarc’h (Rennes), L. Heyries (Marseille), P. Hillon (Dijon), I. Hubert (Angers), D. Larrey (Montpellier), B. Lesgourgues (Montfermeil), E.H. Metman (Tours), J.P. Miguet and S. Bresson-Hadni (Besançon), F. Mion (Lyon), J. Moreau (Toulouse), X. Moreau (Ollioules), D. Ouzan (Saint Laurent du Var), A. Pariente (Pau), J.C. Paris (Lille), J.P. Pascal (Toulouse), S. Pol (Paris), R. Poupon (Paris), T. Poynard and V. Ratziu (Paris), J.J. Raabe (Metz), D. Ribard (Nîmes), D. Roulot (Bobigny), D. Samuel (Villejuif), P. Sogni (Paris), A. Tran (Nice), C. Trépo and F. Zoulim (Lyon), D. Vetter (Strasbourg), C. Wartelle Bladou (Aix-en-Provence), J.P. Zarski (Grenoble)
Funding for the Lamivir study was provided by Laboratoire GlaxoSmithKline, France |
1. Ganem D, Prince AM. Hepatitis B virus infection--natural history and clinical consequences. N Engl J Med. 2004;350:1118–1129. 2. Nowak MA, Bonhoeffer S, Hill AM, Boehme R, Thomas HC, McDade H. Viral dynamics in hepatitis B virus infection. Proc Natl Acad Sci U S A. 1996;30(93):4398–4402. 3. Lai CL, Chien RN, Leung NW, Chang TT, Guan R, Tai DI, Ng KY, Wu PC, Dent JC, Barber J, Stephenson SL, Gray DF. A one-year trial of lamivudine for chronic hepatitis B. Asia Hepatitis Lamivudine Study Group N Engl J Med. 1998;339:61–68. 4. Dienstag JL, Schiff ER, Wright TL, Perrillo RP, Hann HW, Goodman Z, Crowther L, Condreay LD, Woessner M, Rubin M, Brown NA. Lamivudine as initial treatment for chronic hepatitis B in the United States. N Engl J Med. 1999;341:1256–1263. 5. Schiff ER, Dienstag JL, Karayalcin S, Grimm IS, Perrillo RP, Husa P, de Man RA, Goodman Z, Condreay LD, Crowther LM, Woessner MA, McPhillips PJ, Brown NA. International Lamivudine Investigator Group. Lamivudine and 24 weeks of lamivudine/interferon combination therapy for hepatitis B e antigen-positive chronic hepatitis B in interferon nonresponders. J Hepatol. 2003;38:818–826. 6. Liaw YF, Leung NW, Chang TT, Guan R, Tai DI, Ng KY, Chien RN, Dent J, Roman L, Edmundson S, Lai CL. Effects of extended lamivudine therapy in Asian subjects with chronic hepatitis B. Asia Hepatitis Lamivudine Study Group. Gastroenterology. 2000;119:172–180. 7. Leung NW, Lai CL, Chang TT, Guan R, Lee CM, Ng KY, Lim SG, Wu PC, Dent JC, Edmundson S, Condreay LD, Chien RN. On behalf of the Asia Hepatitis Lamivudine Study Group. Extended lamivudine treatment in patients with chronic hepatitis B enhances hepatitis B e antigen seroconversion rates: results after 3 years of therapy. Hepatology. 2001;33:1527–1532. 8. Tassopoulos NC, Volpes R, Pastore G, Heathcote J, Buti M, Goldin RD, Hawley S, Barber J, Condreay L, Gray DF. Efficacy of lamivudine in subjects with hepatitis B e antigen-negative/hepatitis B virus DNA-positive (precore mutant) chronic hepatitis B. Hepatology. 1999;29:889–896. 9. Honkoop P, Niesters HG, de Man RA, Osterhaus AD, Schalm SW. Lamivudine resistance in immunocompetent chronic hepatitis B. Incidence and patterns J Hepatol. 1997;26:1393–1395. 10. Allen MI, Deslauriers M, Andrews CW, Tipples GA, Walters KA, Tyrrell DL, Brown N, Condreay LD. Identification and characterization of mutations in hepatitis B virus resistant to lamivudine. Lamivudine Clinical Investigation Group. Hepatology. 1998;27:1670–1677. 11. Gauthier J, Bourne EJ, Lutz MW, Crowther LM, Dienstag JL, Brown NA, Condreay LD. Quantitation of hepatitis B viremia and emergence of YMDD variants in subjects with chronic hepatitis B treated with lamivudine. J Infect Dis. 1999;180:1757–1762. 12. Tipples GA, Ma MM, Fischer KP, Bain VG, Kneteman NM, Tyrrell DL. Mutation in HBV RNA-dependent DNA polymerase confers resistance to lamivudine in vivo. Hepatology. 1996;24:714–717. 13. Ling R, Harrison TJ. Functional analysis of mutations conferring lamivudine resistance on hepatitis B virus. J Gen Virol. 1999;80:601–606. 14. Lai CL, Dienstag J, Schiff E, Leung NW, Atkins M, Hunt C, Brown N, Woessner M, Boehme R, Condreay L. Prevalence and clinical correlates of YMDD variants during lamivudine therapy for subjects with chronic hepatitis B. Clin Infect Dis. 2003;36:687–696. 15. Liaw YF, Chien RN, Yeh CT, Tsai SI, Chu CM. Acute exacerbation and hepatitis B virus clearance after emergence of YMDD motif mutation during lamivudine therapy. Hepatology. 2000;119:172–180. 16. Rizzetto M, Tassopoulos NC, Goldin RD, Esteban R, Santantonio T, Heathcote EJ, Lagget M, Taak NK, Woessner MA, Gardner SD. Extended lamivudine treatment in patients with HBeAg-negative chronic hepatitis B. J Hepatol. 2005;42:173–179. 17. Chang TT, Lai CL, Chien RN, Guan R, Lim SG, Lee CM, Ng KY, Nicholls GJ, Dent JC, Leung NW. Four years of lamivudine treatment in Chinese patients with chronic hepatitis B. J Gastroenterol Hepatol. 2004 Nov;19(11):1276–82. 18. Liaw YF, Sung JJ, Chow WC, Farrell G, Lee CZ, Yuen H, Tanwandee T, Tao QM, Shue K, Keene ON, Dixon JS, Gray DF, Sabbat J. Cirrhosis Asian Lamivudine Multicentre Study Group. Lamivudine for patients with chronic hepatitis B and advanced liver disease. N Engl J Med. 2004 Oct 7;351(15):1521–31. 19. Di Marco V, Marzano A, Lampertico P, Andreone P, Santantonio T, Almasio PL, Rizzetto M, Craxi A. Italian Association for the Study of the Liver (AISF) Lamivudine Study Group, Italy. Clinical outcome of HBeAg-negative chronic hepatitis B in relation to virological response to lamivudine. Hepatology. 2004;40:883–891. 20. Paik YH, Chung HY, Ryu WS, Lee KS, Lee JS, Kim JH, Lee CK, Chon CY, Moon YM, Han KH. Emergence of YMDD motif mutant of hepatitis B virus during short-term lamivudine therapy in South Korea. J Hepatol. 2001;35:92–98. 21. Da Silva LC, da Fonseca LE, Carrilho FJ, Alves VA, Sitnik R, Pinho JR. Predictive factors for response to lamivudine in chronic hepatitis B. Rev Inst Med Trop Sao Paulo. 2000;42:189–196. 22. Wakil SM, Kazim SN, Khan LA, Raisuddin S, Parvez MK, Guptan RC, Thakur V, Hasnain SE, Sarin SK. Prevalence and profile of mutations associated with lamivudine therapy in Indian subjects with chronic hepatitis B in the surface and polymerase genes of hepatitis B virus. J Med Virol. 2002;68:311–318. 23. Knodell RG, Ishak KG, Black WC, Chen TS, Craig R, Kaplowitz N, Kiernan TW, Wollman J. Formulation and application of a numerical scoring system for assessing histological activity in asymptomatic chronic active hepatitis. Hepatology. 1981;1:431–435. 24. Bedossa P, Poynard T. An algorithm for the grading of activity in chronic hepatitis C. The METAVIR Cooperative Study Group. Hepatology. 1996;24:289–293. 25. Grandjacques C, Pradat P, Stuyver L, Chevallier M, Chevallier P, Pichoud C, Maisonnas M, Trepo C, Zoulim F. Rapid detection of genotypes and mutations in the pre-core promoter and the pre-core region of hepatitis B virus genome: correlation with viral persistence and disease severity. J Hepatol. 2000;33:430–439. 26. Chu CJ, Keeffe EB, Han SH, Perrillo RP, Min AD, Soldevila-Pico C, et al. Prevalence of HBV precore/core promoter variants in the United States. Hepatology. 2003;38(3):619–28. 27. Lindh M, Hannoun C, Dhillon AP, Norkrans G, Horal P. Core promoter mutations and genotypes in relation to viral replication and liver damage in East Asian hepatitis B virus carriers. J Infect Dis. 1999;179(4):775–82. 28. Parekh S, Zoulim F, Ahn SH, Tsai A, Li J, Kawai S, et al. Genome replication, virion secretion, and e antigen expression of naturally occurring hepatitis B virus core promoter mutants. J Virol. 2003;77(12):6601–12. 29. Zoulim F. Mechanism of viral persistence and resistance to nucleoside and nucleotide analogs in chronic hepatitis B virus infection. Antiviral Res. 2004;64:1–15. 30. Locarnini S, Hatzakis A, Heathcote J, Keeffe EB, Liang TJ, Mutimer D, Pawlotsky JM, Zoulim F. Management of antiviral resistance in patients with chronic hepatitis B. Antivir Ther. 2004;9:679–693. 31. Benhamou Y, Bochet M, Thibault V, Di Martino V, Caumes E, Bricaire F, Opolon P, Katlama C, Poynard T. Long-term incidence of hepatitis B virus resistance to lamivudine inhuman immunodeficiency virus-infected subjects. Hepatology. 1999;30:1302–1306. 32. Lau DT, Khokhar MF, Doo E, Ghany MG, Herion D, Park Y, Kleiner DE, Schmid P, Condreay LD, Gauthier J, Kuhns MC, Liang TJ, Hoofnagle JH. Long-term therapy of chronic hepatitis B with lamivudine. Hepatology. 2000;32:828–834. 33. Nafa S, Ahmed S, Tavan D, Pichoud C, Berby F, Stuyver L, Johnson M, Merle P, Abidi H, Trepo C, Zoulim F. Early detection of viral resistance by determination of hepatitis B virus polymerase mutations in subjects treated by lamivudine for chronic hepatitis B. Hepatology. 2000;32:1078–1088. 34. Zöllner B, Petersen J, Schroter M, Laufs R, Schoder V, Feucht HH. 20-fold increase in risk of lamivudine resistance in hepatitis B virus subtype adw. Lancet. 2001;357:934–935. 35. Akuta N, Suzuki F, Kobayashi M, Tsubota A, Suzuki Y, Hosaka T, Someya T, Kobayashi M, Saitoh S, Arase Y, Ikeda K, Kumada H. The influence of hepatitis B virus genotype on the development of lamivudine resistance during long-term treatment. J Hepatol. 2003;38:315–321. 36. Suzuki Y, Arase Y, Ikeda K, Saitoh S, Tsubota A, Suzuki F, Kobayashi M, Akuta N, Someya T, Miyakawa Y, Kumada H. Histological improvements after a three-year lamivudine therapy in subjects with chronic hepatitis B in whom YMDD mutants did not or did develop. Intervirology. 2003;46:164–170. 37. Dienstag JL, Goldin RD, Heathcote EJ, Hann HW, Woessner M, Stephenson SL, Gardner S, Gray DF, Schiff ER. Histological outcome during long-term lamivudine therapy. Gastroenterology. 2003;124:105–117. 38. Hadziyannis SJ, Papatheodoridis GV, Dimou E, Laras A, Papaioannou C. Efficacy of long-term lamivudine monotherapy in patients with hepatitis B e antigen-negative chronic hepatitis B. Hepatology. 2000;32:847–851. 39. Lok AS, Lai CL, Leung N, Yao GB, Cui ZY, Schiff ER, Dienstag JL, Heathcote EJ, Little NR, Griffiths DA, Gardner SD, Castiglia M. Long-term safety of lamivudine treatment in patients with chronic hepatitis B. Gastroenterology. 2003;125:1714–1722. 40. Myers RP, Tainturier MH, Ratziu V, Piton A, Thibault V, Imbert-Bismut F, Messous D, Charlotte F, Di Martino V, Benhamou Y, Poynard T. Prediction of liver histological lesions with biochemical markers in patients with chronic hepatitis B. J Hepatol. 2003;39:222–30. 41. Poynard T, Zoulim F, Ratziu V, Degos F, Imbert-Bismut F, Deny P, Landais P, EI Hasnaoui A, Slama A, Blin P, Thibault V, Parvaz P, Munteanu M, Trépo C. Longitudinal assessment of histology surrogate markers (Fibrotest-Actitest) during lamivudine therapy in patients with chronic hepatitis B infection. Am J Gastroenterol. in press. |
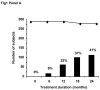 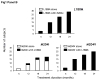 | Figure 1 Emergence of lamivudine-resistant viral strains over the study |
 | Table 1 Demographic and clinical characteristics of the study population. BMI: body mass index; PVHT: portal vein hypertension. |
 | Table 2 Indices of hepatitis B infections in the study population. ALT: alanine aminotransferase; ULN: upper limit of normal. |
 | Table 3Presence of mutations in the YMDD motif of the HBV polymerase gene at study end according to HBe serotype and LiPA genotype at inclusion. The percentages are calculated by column. One subject from whom strains with LiPA genotypes A and D were isolated (more ...) |
 | Table 4 Emergence of lamivudine-resistant mutations as a function of baseline variables. ND: Not determined due to low subject numbers. |
|

